26/02/2025
Almost a decade ago, a Personalised Medicine Strategy Board was established by NHS England, with the purpose of developing a strategy to “better manage patients’ health and to target therapies to achieve the best outcomes in the management of a patient’s disease or predisposition to disease” [1].
Personalised medicine refers to treatments and other medical interventions which are tailored to the specifics of an individual. This personalisation can be made possible by analysis of the individual’s genome. Patterns in disease and responses to certain treatments can be correlated with genetic markers, allowing more robust conclusions about a diagnosis to be made. Such a concept has the opportunity to move away from the traditional ‘one-size-fits-all’ approach to healthcare, and instead deliver tailored remedies that are not only more effective but also come with reduced risks of adverse reactions and harmful side effects.
Through the NHS Genomic Medicine Service, whole genome sequencing may be made available for certain conditions where the evidence has shown that it can be beneficial – specifically cancers and rare diseases [2]. But there exists significant potential in this approach for treating a much broader range of conditions than these specific examples. However, this potential leap forward in patient outcomes hinges on the ability to accurately and, just as crucially affordably, sequence the individual’s genome.
The sequencing of genomes has been in continuous development for over 50 years, with the goals of reducing sequencing costs and complexity, whilst ensuring fast and accurate reads, ever present during this development.
In 2004, the National Human Genome Research Institute launched a US-government backed programme to support technological development to reduce the cost of genome sequencing. Following this programme, the cost associated with a single whole genome sequencing (WGS) has dropped substantially, from over $10 million to under $1000 by 2022 [3]. This rapid rate of development has surpassed that predicted by Moore’s law by a significant margin.
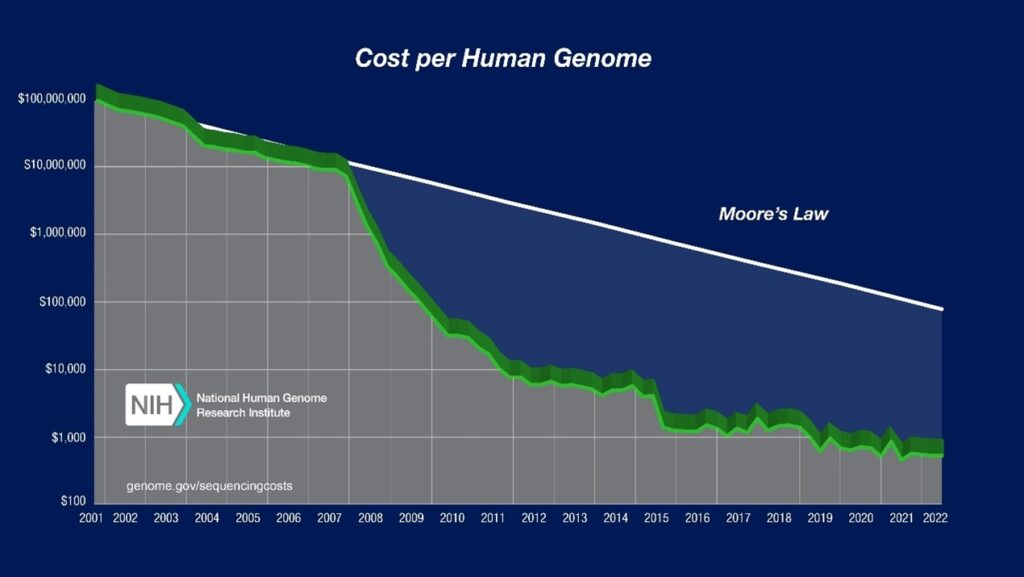
https://www.genome.gov/about-genomics/fact-sheets/Sequencing-Human-Genome-cost
By continuing to reduce the cost of WGS, technologies and developments such as personalised medicine will become more accessible to the public. The next milestone of a sub-$100 WGS will however require further technological development of sequencing techniques.
Nanopore sequencing emerged as a concept in the 1990s, and is a contender in the advancement towards affordable personalised medicine for the masses.
One example of nanopore sequencing involves detecting a current across a nanopore in a membrane as single strands of polynucleotides are threaded therethrough. This technology can be broadly divided into two separate groups: biological nanopore sequencing in which polynucleotides are drawn through a transmembrane protein, and solid-state nanopore sequencing, in which the nanopore is instead formed in a non-biological substrate by physical or chemical means. By applying signal processing methods to the detected current, the bases which make up the polynucleotides can be determined.
Nanopores sequencing not only promises real-time sequencing of longer sequences, but also reduced computational requirements, and in small form-factor devices. This may all contribute to nanopore sequencing leading the way in more affordable WGS, and bringing personalised medicine to the forefront in healthcare.
Patent publications in the nucleic acid sequencing space which mention nanopores have been steadily increasing for the past decade, with last year seeing over 800 patent publications. Interest in the field is therefore steadily but consistently building.
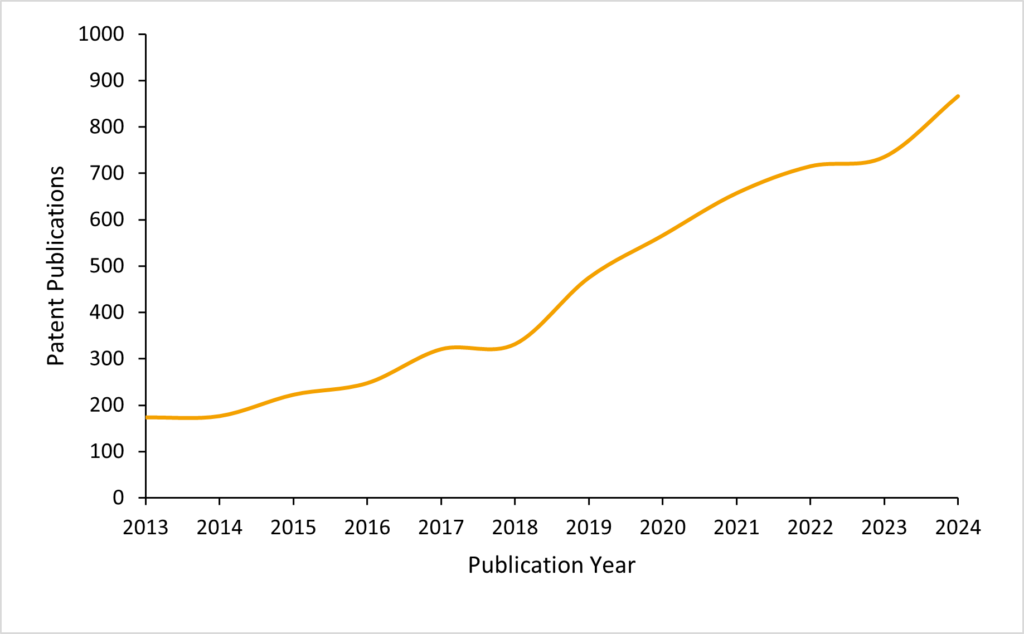
Of the 5,500 publications since 2013, no singular assignee hold a majority share of the filings. Rather, a broad range of companies occupy this patent filing space.
The Swiss multi-national Roche are assignees with largest share of 773 of these publications, representing only 14% of the total number.
Roche’s commitment to this technological direction for sequencing was also made clear in May 2020 with their acquisition of Stratos Genomics [4]. The Seattle-based company aims to develop ultra-low-cost genome sequencing by novel next-generation sequencing chemistry referred to as Sequencing by Expansion (SBX).
A significant issue associated with present nanopore sequencing technologies is the high translocation speed of polynucleotides through nanopores. These speeds mean that several nucleotides can pass through a nanopore in mere microseconds. This results in a low signal-to-noise ratio in the measured current, which reduces sequencing accuracy.
SBX intends to tackle this problem, and is the subject of a number of patents and applications filed by Stratos Genomics, an example of which being published PCT application WO 2020/263703 A1.
The technology works by building a chain from monomeric substrates from a nucleic acid template to be sequenced, as illustrated in Figure 1B below. This is followed by cleavage of the chain at selected locations. This cleavage results in the loops of each monomeric substrate ‘opening up’, forming a linearly elongated molecule – referred to as an “Xpandomer”. This is illustrated in Figure 1C below.
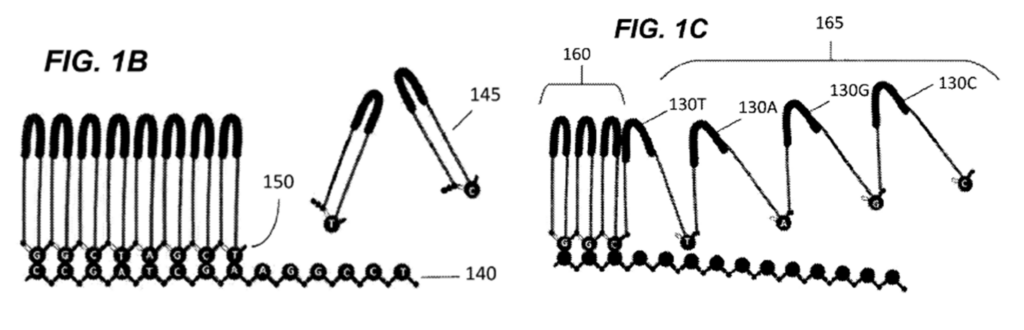
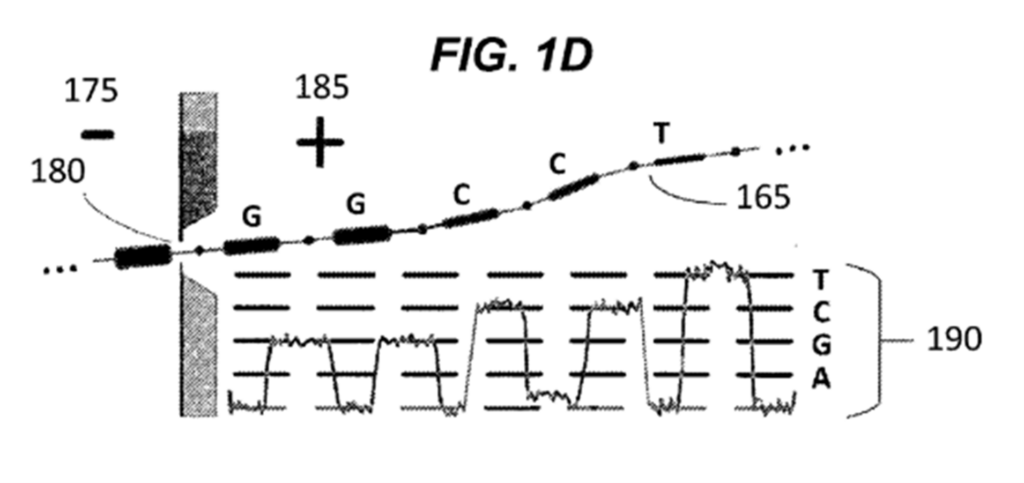
As each monomeric substrate specifically corresponds to each DNA base in the template nucleic acid, reporter constructs denoted by 130 in Figure 1C above are disposed along each monomeric substrate. Each reporter construct is therefore characteristic of the DNA base at that location in the elongated molecule, and the information from the original nucleic acid template is preserved. When the Xpandomer is subsequently drawn through the nanopore, as shown in Figure 1D below, the reporter constructs assist by providing a higher signal-to-noise ratio in the detected current, which increases sequencing accuracy.
Four years on from this acquisition, at their Diagnostics Day in May 2024, Roche announced their intention of launching a nanopore sequencer exploiting this unique SBX technology from 2025 onwards, highlighting it as a key growth opportunity with significant market potential [5]. This anticipated product would include a reported 8 million microwells for sequencing in parallel, further reducing sequencing timescales, enhancing accuracy, and reducing the footprint of the sequencing device.
The technology surrounding nanopore sequencing is therefore clearly still developing, and each step draws personalised healthcare further into cost-viability. As sequencing costs continue to fall, it will be interesting to see if these developments eventually contribute to genetic sequencing becoming routinely considered for the majority of treatments.
If you need assistance or advice to protect your innovations, then please do get in touch with one of our attorneys at Reddie & Grose.
This article is for general information only. Its content is not a statement of the law on any subject and does not constitute advice. Please contact Reddie & Grose LLP for advice before taking any action in reliance on it.



